Determinants of miRNA-target interaction & Circular RNA
Team: Stanislas Werfel, Sabine Brummer, Saskia Rausch
How can targets of a microRNA be identified? And why are some targets strongly regulated while others are almost not regulated at all? Although both questions are as old as the knowledge of the existence of microRNAs as gene regulators, they are still unresolved and need to be further elucidated before we can fully appreciate the function and relevance of a microRNA.
Several methods for the identification of potential targets have been established at our institute. Among them, next generation RNA sequencing allows to make transcriptome-wide predictions of the overall targetome of a microRNA and automated reporter assays allow for confirmation of selected targets.
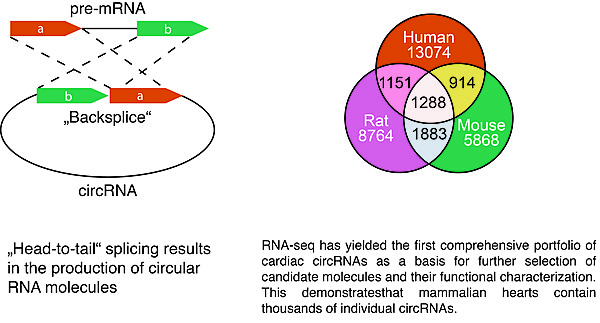
The discovery of circular RNAs or circRNAs has recently sparked a whole new field of non-coding RNA research. Our institute was among the first groups to categorize and characterize circRNAs present in the human heart in health and disease (Werfel et al., J. Mol. Cell. Cardiol., 2016). We thereby could observe that also a large number of circular RNAs are conserved among the species and most interestingly that some important genes, such as titin or the ryanodine receptor produce a multitude of different circRNAs, in the order of dozens to hundreds. Elucidating the function of circular RNAs might prove an important contribution to understanding the processes underlying the proper function of the heart muscle.
References:
S. Werfel, S. et al., J. Mol. Cell. Cardiol. 98. 2016.